Release of GWAS Data of Foxtail Millet
Foxtail millet (Setaria italica) is an important grain crop world-widely grown in arid regions. However, the genetic basis of most agronomic traits and genetic diversity in foxtail millet are still poorly understood. Researchers from Chinese Academy of Sciences reported the construction of a haplotype map and large-scale GWAS in foxtail millet.
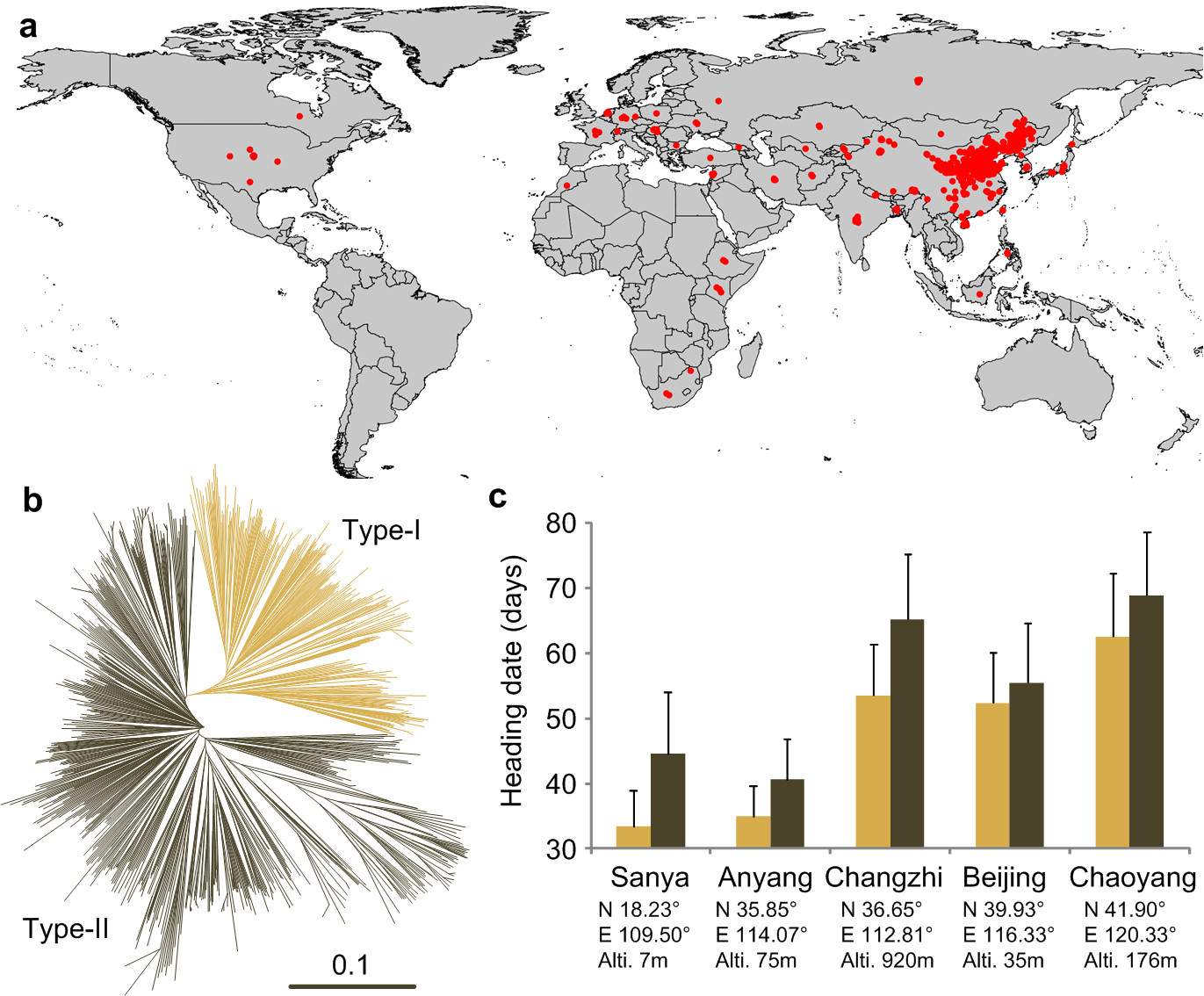
In a recent work, 916 diverse foxtail millet varieties including both traditional landraces and modern genetic-improved cultivars to detect genomic variations were sequenced. A total of 2.58 million SNPs were identified, and 0.8 million common SNPs were used to construct a haplotype map of the foxtail millet genome. On the basis of the neighbor-joining tree, the foxtail millet varieties were classified into two divergent groups that were strongly correlated with early and late flowing times. The 916 varieties were phenotyped under five different environments, and totally 512 loci associated with 116 agronomic traits were identified through genome-wide association study. De novo assembly of deeply sequenced genomes of a Setaria viridis accession (the wild progenitor of S. italica) and a S. italica variety were also performed with most complex inter-species and intra-species variants identified. In the search for signatures of selection, 36 selective sweeps during modern breeding were further identified.
This study provides fundamental resources and platform for genetics research and genetic improvement in foxtail millet. The work entitled “A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica)” has been published in Nature Genetics in Jun 24.
Geographic distribution and genetic structures of 916 foxtail millet varieties. (Image by Dr. Bin Han’s Group)
CONTACT:
Bin Han
National Center for Gene Research
Institute of Plant Physiology and Ecology,Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Tel: 86-21-64845260; Email: bhan@ncgr.ac.cn