Scientists Reveal Interaction between Two Plant Timing MicroRNAs
In Arabidopsis thaliana, the spatial pattern of trichome is a hallmark of phase transition and governed by miR156-targeted SPLs, but how the timely increasing SPL output is modulated remains elusive. Recently, researchers from the Chinese Academy of Sciences (CAS) report that another conserved timing MicroRNAs, miR171, affects the SPL activity through antagonistic interaction between target proteins.
XUE Xueyi, a Ph.D. graduate in the research group led by Dr. CHEN Xiaoya from the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, CAS, used stem trichome as screen system to investigate the interaction of miR156-SPL module with other developmental or environmental signaling pathways. Through a series of genetic experiments, XUE et al. found that a group of GRAS family members, LOST MERISTEMS 1 (LOM1), LOM2 and LOM3, targeted by miR171, promote trichome formation via SPLs. Further analysis revealed that physical interaction between the N-terminus of LOMs and SPLs underpins the repression of SPL activity. Furthermore, they provide evidence that MIR171A gene expression is regulated by its targeted LOMs, forming a homeostatic feedback loop.
This research establishes an age-dependent regulatory network composed of two timing miRNAs, which sheds light on how plants coordinate environmental cues with endogenous flowering pathway.
This work entitled “Interaction between Two Timing MicroRNAs Controls Trichome Distribution in Arabidopsis” was published in PLoS Genetics on April 3, 2014. This research was supported by grants from the State Key Basic Research Program of China (2010CB126004) and Chinese Academy of Sciences (KSCX2-YW-N-057).
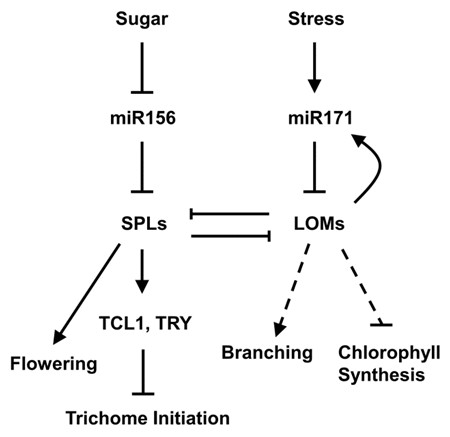
miR171-LOM and miR156-SPL interaction in regulating trichome formation (Image by Dr. CHEN Xiaoya's group)
