Unveiling the mechanism of oosporein biosynthesis and its effect on immune inhibition in insects
Quinones are widely distributed in nature and exhibit diverse biological or pharmacological activities; however, their biosynthetic machineries are largely unknown. Oosporein, a red 1,4-bibenzoquinone derivative was first identified from the ascomycete insect pathogen Beauveria bassiana in the 1960s and exhibits antibiotic, antiviral, antifungal and insecticidal activities.
After finishing the genomic study of
B. bassiana,
Prof. WANG Chengshu’s group at the Institute of Plant and Physiology and Ecology, Shanghai Institutes for Biological Sciences, CAS, unveiled the biosynthetic machinery of oosporein in fungi. The polyketide synthase (PKS) gene cluster includes seven genes for oosporein biosynthesis. The PKS OpS1 produces orsellinic acid that is hydroxylated to 6-methyl-benzenetriol by the hydroxylase OpS4. The intermediate is oxidized to 6-methyl-benzenetetrol by the cupin-family oxygenase OpS7. The latter is further dimerized to oosporein by the catalase OpS5. The transcription factor OpS3 regulates intrapathway gene expression. Due to the keto-enol tautomerism, different intermediate metabolites were also identified, including the previously unidentified 5,5’-dideoxy-oosporein. Insect bioassays revealed that oosporein is employed by the fungus to facilitate insect killing by evading host cellular and humoral immunities. The results advance the current understanding of quinone biosynthetic mechanism, and the small-molecule type effector deployed by fungi to interact with their hosts.
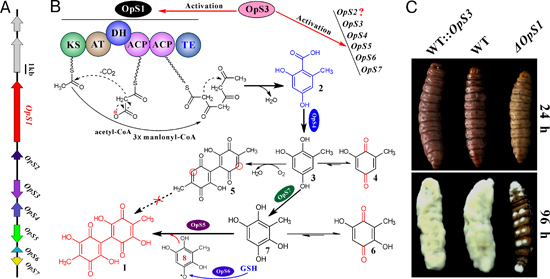
A, Gene cluster for oosporein biosynthesis; B, Oosporein biosynthetic pathway; C, Insect bioassays showing the delay in mycosis of insects after the loss of oosporein-producing ability in
B. bassiana.
