Construction of polyketide overproducing Escherichia coli strains via synthetic antisense RNAs
Identification of genes that affect the product accumulation phenotype of recombinant strains is always an important problem in industrial strain construction. Rapid assessment and optimization of the incompatibility between the exogenous biosynthetic modules and the host metabolic modules remains a challenge for heterologous biosynthesis in the context of synthetic biology.
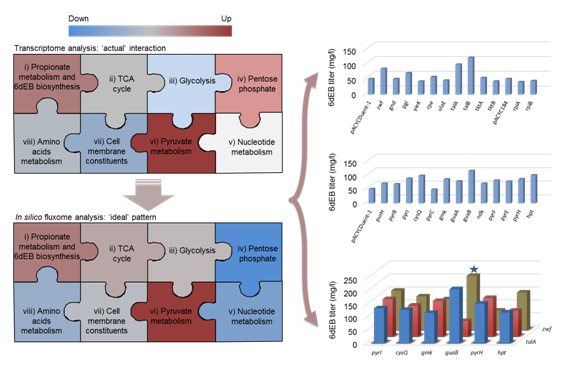
The research group led by Prof. Yong Wang at Institute of Plant Physiology and Ecology (SIPPE), Shanghai Institutes for Biological Sciences (SIBS), CAS, developed a systematic approach based on in silico genome-scale metabolic flux analysis and comparative transcriptome profiling to characterize the module interactions and applied it to illustrate the problematic interactions between foreign erythromycin precursor 6-deoxyerythronolide B (6dEB) module and the native metabolic modules in E. coli. In comparison with the in silico ‘ideal situation’ for the maximum rate of 6dEB biosynthesis, the ‘actual situation’ represented by transcriptome profiling showed tremendous differences in the overall trends of flux changes of metabolic modules, especially for the pentose phosphate pathway and the nucleotide metabolism module. By analyzing these discrepancies, potential targets were firstly identified for improving 6dEB biosynthesis. Cells with engineered 18 targets showed more than 20% increase in 6dEB yield, and combinatorial repression of targets in these two modules resulted in >60% increase in 6dEB titer, e.g., anti-guaB and anti-zwf led to a 296.2% increase in 6dEB production (210.4 mg/l in flask). This is the highest yield yet reported for polyketide heterologous biosynthesis in E. coli. These results, which improving the interactions and adaptations within and among the modules to meet with those of the theoretical ’ideal‘ situations at systemic levels, would be an effective strategy to improve the yield of heterologous products in the chassis cell. The results reported here are of significant improvement over previous work.
This study entitled ’Construction of polyketide overproducing Escherichia coli strains via synthetic antisense RNAs based on in silico fluxome analysis and comparative transcriptome analysis‘ has been published on line on Biotechnology Journal ( http://onlinelibrary.wiley.com/doi/10.1002/biot.201500351/abstract).
The work was funded by Hundred-Talent Program (Chinese Academy of Sciences), the National Basic Research Program of China (973 Program), the National Natural Science Foundation of China.
CONTACT:
Prof. Yong Wang, PhD, Principal Investigator, Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China. Tel/Fax: 54924295; E-mail:
yongwang@sibs.ac.cnImproving pathway imbalance (or ill interac- tion) can highly enhance downstream product biosynthesis in heterologous host. Metabolic pathways were abstracted into eight major functional modules, and cellular module inter- action was analyzed for 6-deoxyerythronolide B (6dEB) production in E. coli based on in silico fluxome analysis and comparative tran- scriptome analysis. The predicted targets were firstly tested for improving the 6dEB production via synthetic antisense RNAs, which led to a highest yield (210.4 mg/L in flask) yet reported for polyketide heterologous biosynthesis in this strain.